Camille Ameline, PhD.
I am an evolutionary biologist with a keen interest in host-microorganisms interactions. After a Masters in Ecology at the University of Rennes and a PhD in Evolutionary Biology at the University of Basel, I gained experience in varied fields of Biology, from Population Genetics to Microbiology. During my PhD I investigated the evolution and genetic architecture of bacterial resistance in a natural population of a crustacean host, Daphnia magna. Throughout my studies and research I have worked - in the field or in the lab - with tits, marmots, spiders, crustaceans and now mice. I enjoy challenging field work as much as tidy lab work. In my current research as a postdoc in the Evolutionary Biology group at the Instituto Gulbenkian de Ciência, I am investigating the interplay between the evolution of the commensal bacterium Escherichia coli in mice gut and the adaptive immune system of the host.

Current Research
How does the immune status of the host influence the evolution of microbiota in the gut?
Gut microbiota is hypothesized to be a major driver of evolution in vertebrates. The diversity and evolution of gut microbiota have been shown to influence speciation, development, and immunity in the host. Because the microbial community in the gut is complex, it remains challenging to study the influence of host traits on the diversity and evolution of gut microbiota.
The evolutionary dynamics of commensal bacteria during colonization of the gut is of particular interest in the context of immune and inflammatory bowel diseases (IBD). In such cases, the natural balance of the gut microbiota is disrupted, and specific species dominate the gut ecosystem. While the composition of the gut ecosystem has been much characterized, little is known about the process of evolution that occurs within each species. I am using the host–bacterium model system Mus musculus–Escherichia coli, as a first step to unravel the emergence of intra-species diversity and the dynamics of mutations in the bacteria while colonizing the host gut.
I am conducting experimental evolution of E. coli in immune-deprived and Inflammatory Bowel Disease (IBD) mutant mice in germ-free conditions, to remove the environmental factor of the surrounding microbiota (Figure 1). Using fluorescently marked strains of E. coli, I will track major evolutionary changes, and study the biogeography of evolution in the intestinal tract after thousands of generations. I will investigate the tempo and mode of E. coli evolution, measuring rates of mutation accumulation and horizontal gene transfer, and determining evolution predictability across hosts with distinct immune status. I will test if the rate of spontaneous mutations is increased in immune-deprived animals; identify the targets of selection using genomic tools; and quantify the strength of selection in follow-up competition experiments.

Figure 1
Previous Research
Microorganisms, such as parasites and gut microbiota, are believed to be major drivers of the evolution of host immunity, and of the maintenance of diversity and sexual reproduction However, the link between theory, observations in natural environments, and underlying genetics is often missing.
In my PhD, I investigated the evolution of resistance in a population of the cyclic parthenogen Daphnia magna that undergoes strong epidemics caused by the bacteria Pasteuria ramosa. This population responds to the epidemics with a highly repeatable pattern of adaptive and rapid increase in resistance (Figure 2). We further found that resistance to the parasite is based of multiple loci, dominance and strong epistasis, providing functional support to the importance of epistasis in host-parasite interactions.
I then investigated the effect of recombination on the evolution of resistance in the host. I used long-term monitoring of the D. magna population and the remarkable capacity of cyclic parthenogens to alternate between clonal and sexual reproduction. In this study we provide empirical evidence for genetic slippage in response to sex in a cyclical parthenogen.
A key aspect of host-parasite interactions is a high specificity between host and parasite genotypes. We described new sites and patterns of attachment of the bacterial spores on the host cuticle (Figure 3), an infection step that has been shown to be highly specific in the D. magna-P. ramosa system.
As a research assistant at the Bee Research Center in the Agroscope, Switzerland, I investigated the influence of paternal genotype on the susceptibility of honey bee to the pathogenic bacterium Melissococcus plutonius, the causative agent of an economically important disease, the European Foulbrood (Figure 4). Using experimentally infected larvae, we showed that susceptibility to the pathogen is modulated by paternal genotype.

Figure 2
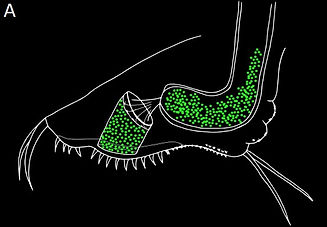
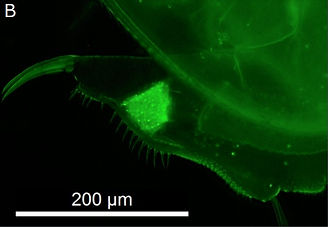
Figure 3
